Part 6 - PCR Detection of Beer Spoilage Explained
November 25, 2020
PCR TECHNOLOGY FOR THE DETECTION OF MICROBIAL CONTAMINANTS EXPLAINED
All beer spoiler microorganisms possess DNA (Deoxyribonucleic acid) which contains the genetic instructions for the development and function of living things. While in animals, plants and yeasts the DNA is contained in the cell nucleus, in bacteria the DNA is present in their cytoplasm (the material within a cell, enclosed by the cell membrane).
Polymerase chain reaction (PCR) is a simple and elegant test that was invented in 1983 by the American biochemist Kary Mullis. As she outlined in her publication (1990), it “lets you pick the piece of DNA you’re interested in and have as much of it as you want. Only trace amounts of DNA are needed for PCR to generate enough copies to be analysed using conventional laboratory methods. For this reason, PCR is a sensitive assay.
Today it is a method used widely in molecular biology and whereas 5 years ago, it was still very much considered a technology that could only be found in the large central laboratories and research institutes due to the investment necessary both in the equipment and the specific sterile laboratory conditions, in more recent times there has been a simplification of the PCR protocols and equipment (thermocyler) thus allowing a “democratisation” of PCR technology so as to be used as a real production tool on site at the brewery and throughout the food production chain.
WHAT IS DNA MADE UP OF?
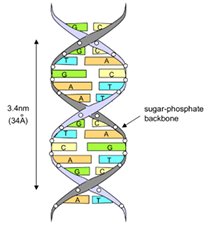
The information in DNA is stored as a code made up of four chemical bases: adenine (A), guanine (G), cytosine (C), and thymine (T). DNA bases pair up with each other, A with T and C with G, to form units called base pairs. Each base is also attached to a sugar molecule and a phosphate molecule. Together, a base, sugar, and phosphate are called a nucleotide. Nucleotides are arranged in two long strands that form a spiral called a double helix.
An important property of DNA is that it can replicate, or make copies of itself. Each strand of DNA in the double helix can serve as a pattern for duplicating the sequence of bases.
A STEP-BY-STEP LOOK AT HOW DOES PCR WORK ON THE DETECTION OF BEER SPOILER MICROBES?
1. Liberate the DNA from bacteria or yeast
Sample is centrifuged to concentrate “beer spoiler cells” before adding a buffer to breakdown cell wall
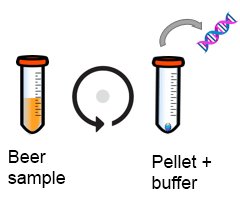
Extraction of beer spoiler DNA from beer
2. Beer spoiler DNA is then added to PCR “master mix solution” which contains all the necessary elements for the reaction (replication of the DNA) and is placed in a Thermocycler.
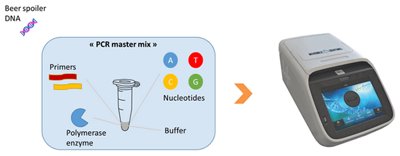
3. A high temperature is applied to separate the 2 strands of DNA (Denaturation) and then reducing the temperature enables the “Beer spoiler specific primers” to seek out their corresponding DNA code and line up opposite them (Annealing).
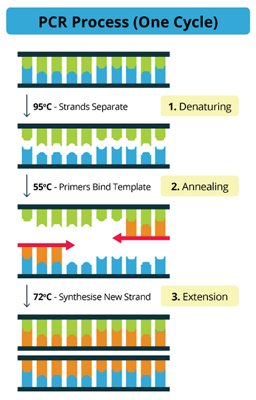
4. With the primers are in position, the enzyme (Polymerase enzyme) knows where to start and finish its synthesis of a new strand to create an new double strand (Extension).
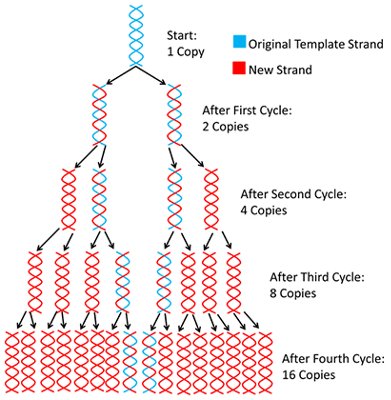
And so 1 strand becomes 2 after one full cycle. The cycles are then repeated in the “Thermo – cycler”
At the end of each PCR cycle, the PCR product or amplicon will increase exponentially because the newly synthesized DNA sequences can be used as templates (in addition to the original DNA template). Usually, 20 to 30 standard PCR cycles are enough to promote an increment of 106 to 109 of the DNA fragments.
5. The reading of the PCR synthesised amplicons can happen in 2 ways!
a) Real time PCR ( qRT-PCR) is based on the principle that higher or lower initial amounts of a specific DNA sequence (beer spoiler presence) will lead to higher or lower concentrations of amplicons respectively. As the amplicons are synthesised, they are tagged with a fluorescence marker that can then be “read” by a laser or optic reader and interpreted according to the thermocycler software to infer the amplicon’s concentration.
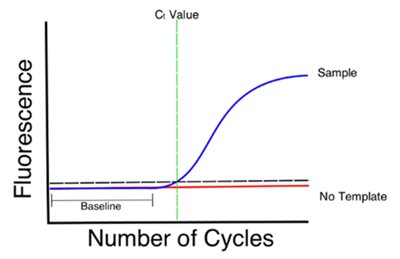
The result is often expressed as a Ct value which is the PCR cycle number at which your sample’s reaction curve intersects a pre-established threshold line. This value tells how many cycles it took to detect a real signal from your samples. Interpretation of the results can often by difficult and requires experience to reliably say if there is a “beer spoiler” presence risk or not.
b) End-stage PCR with Rapid test reading is also, as for qRT-PCR based on the principle that higher or lower initial amounts of a specific DNA sequence (beer spoiler presence) will lead to higher or lower concentrations of amplicons respectively. The difference is that a fixed number of cyclers are performed and as each amplicon is synthesised, instead of attaching a fluorescence marker to it, a biomarker is attached which will then be detected by a specific “antibody” that is present on a rapid test. As for a “pregnancy test”, the result is visual with not only presence / absence but also when present, the degree of line intensity can be measured against a “standard” scorecard to give the semi-quantitative level of “beer spoiler cells” that were originally present.
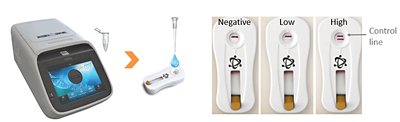
PCR - Rapid test result interpretation
PASTEURISATION OF BEER AND SUBSEQUENT CONTROL FOR BEER SPOILER PRESENCE USING PCR
While as mentioned above, PCR technology is rapid, sensitive and specific making it an ideal tool for the brewery laboratory, it is not able to differentiate DNA coming from live cells versus those from dead ones. As far as PCR is concerned, DNA is DNA. For many craft breweries this is not an issue however for those craft and industrial breweries who pasteurise their beer the use of PCR post pasteurisation (i.e. prior to bottling or on the finished product) can leave the QC managers scratching their heads when they have a positive “hit” on a sample.
Is it from live cells that have survived the pasteurisation and thus possess a risk to the quality of the finished product or is it simply the remnant DNA after the cell has been killed and although it alerts to QC manager to an upstream issue, it no longer poses a threat to the quality of the beer during its shelf life.
In some cases, the PCR manufacturer will propose a “post pasteurisation” enrichment step of at least 24 hours prior to analysis so that should dead cell DNA be present, it will be diluted out during the enrichment and will obviously not increase during enrichment. The PCR result post enrichment depends nevertheless on the level of contamination prior to pasteurisation and time that the liberated DNA remains intact and this will depend on many different beer variables (e.g. pH, temperature …). So called “false” positives are still possible.
Another approach is to expose the DNA to a molecule (e.g. PMAxx TM - Biotium, Inc.) which has a high affinity for DNA and binds with it but is unable to cross the viable cell to access the DNA within prior to amplification. Once bound and exposed to a specific light wavelength, the “fixed” DNA is unable to be multiplied during the amplification step of the PCR. The end result is that should a positive “hit” be observed, you know it is coming from live and viable cells!
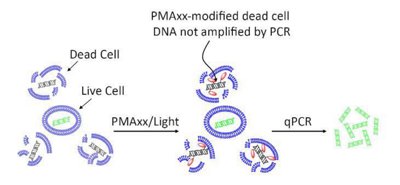
KEY POINTS
• Polymerase Chain Reaction (PCR) technology by its virtue of detecting specific DNA code sequences for the beer spoilers of interest is both very specific and sensitive
• Over the past 5 years, PCR has undergone a democratisation as a result of the protocol simplification and reduction in the capital outlay for the equipment.
• Today, PCR is considered a truly “production orientated” tool because of the same day result capability
• There are 2 approaches to PCR technology (1) End Stage PCR : amplification of the selected DNA segment for a fixed number of cycles and then analysis of the result (2) RT-PCR : during the amplification process, analysis in Real Time after each cycle of the DNA that is present
• PCR enables both a qualitative and quantitative results according to the platform
• QC of pasteurised beers using PCR requires an additional step to avoid the possibility of “false positives” for viable cells