BIOMÉRIEUX EPISEQ®
Next Generation Sequencing (NGS) Data Analysis Platform
Provides easy-to-use applications for clinical microbiologists: bacterial and fungal identification, clinical pathogen outbreak management, microbiome profiling, and SARS-CoV-2 genomics variants identification.
Disclaimer: Product availability varies by country. Please consult your local bioMérieux representative for product availability in your country.

- EPISEQ ID
- EPISEQ CS
- EPISEQ 16S
- EPISEQ SARS-COV-2
- Resources
EPISEQ® ID
RESEARCH USE ONLY
For Bacterial and Fungal Identification
EPISEQ® ID is an easy-to-use web-based application for the identification of bacteria and fungi through their respective 16S and ITS broad-range PCR sequences.
The application is powered by Oxford Nanopore Technologies (ONT) sequencers data.
Ease of Use:
- An intuitive and easy solution to process tNGS FASTQ file directly from Oxford Nanopore Technologies (**) sequencer
- Offers search functionalities through stored historical samples
- Produces a validation page for the detected organisms
- Usable by microbiologists without bioinformatic knowledge
- Provides a comprehensive PDF report
Microbiology Expertise to Deliver Reliable and Trustworthy Results
- A curated database leveraging Oxford Nanopore Technologies long reads sequencing, optimized for full length bacterial 16S and fungal ITS+D1D2 regions.
- EPISEQ® ID provides the list of all bacteria and fungi detected in the sequencing data thanks to its comprehensive database:
The knowledge base is built on curated public and privately owned organism databases, combined with bioMérieux microbiological expertise helping to identify more than 3000 bacteria and 1200 fungi, comprehensive coverage for the clinical species of concern.
*(according to WHO, FDA argos and publications) - Accuracy of the result with a Confidence Score:
Users can rely on the list of organisms detected with this indicator rating the relevance of the species identification based on the amount and quality of sequences enriched with a degree of similarity against the reference database.
Fast and Intuitive Workflow
01 – Upload
From your personal account you can directly upload yourOxford Nanopore Technologies FASTQ file.
EPISEQ® ID is optimized for the full 16S region of bacteria and ITS to D1D2 region for fungi.
EPISEQ® ID is also compatible with amplicon on partial 16S and partial ITS and D1D2 genes.
02 – Analyze
In a single click, EPISEQ® ID assigns bacterial and fungal identification.
EPISEQ® ID delivers a comprehensive list of detected bacteria and fungi at the most reliable taxonomical level or to the closest species level in case of upper identification level.
Multiple Quality Controls allow the user to rely on the delivered results.
03 – Validate and Report
Once the analysis is completed, a validation page allows to either approve or reject all detected organisms.
A pdf report is available once the validation is completed.
EPISEQ® CS
NOT FOR DIAGNOSTIC USE
For Clinical Pathogens Outbreak Monitoring
EPISEQ® CS performs bacterial typing based on Whole Genome Sequencing data and Mutli Resistant Bacteria characterization. It helps confirm Healthcare-Associated Infections (HAIs) outbreaks early and accurately. It allows to track level of resistance genes within the hospital. It reports decisive data to further investigate a potential outbreak or cluster, thus triggering infection prevention and control (IPC) protocols at an early stage to limit the spread of the outbreak.
Ease of Use
- A one-click solution for cluster determination and outbreak management
- Compatible with Illumina and Oxford Nanopore Technologies
- Comprehensive sample characterization and genotyping info
- Phylogenetic visualization tools (dendrogram and minimum spanning tree)
- PDF-downloadable reports
Microbiology Expertise / Antimicrobial Resistance
- Covers the vast majority of HAI-related organisms
- Detects and alerts in case of plasmids outbreak when clusters form, even across different species
- Assesses the phylogenetic relevance of your suspected bacterial outbreak against database of up to 80 000 bacterial genomes
- Built-in wgMLST scheme for each organism
- Display of conventional typing data where available (MLST, spa type, serotype, pathotype)
- More than 4,000 resistance genes embedded with associated phenotypes
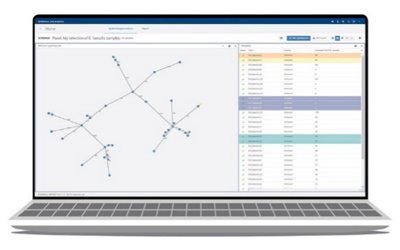
EPISEQ® CS
Fast and Intuitive Workflow
01 – Upload
From your personal account, you can directly upload your Illumina or Oxford Nanopore Technologies sequencing data.
Download the Illumina application note PDF.
(Min. coverage recommended: 45x; Min. sequencing length: 150 PE)
02 – Analyze
In a single click, perform the genomic characterization and a phylogenetic analysis of the strains based on whole genome MLST of the strains. The newly obtained results will be automatically compared to your previous submitted samples.
03 – Report
An easy-to-interpret report is available once the analysis is complete. You now have decisive data to further investigate a potential outbreak or cluster, including resistome, virulome, and plasmid information.
The report provides the most precise information on both sample and group level to address your needs during an outbreak situation:
- Confirms the strain identification
- Determines genetic relatedness among your own strains and the strains in the knowledge base
- Provides resistome, virulome, and plasmid information
- Identifies and differentiates outbreak strains from endemic strains
Extensive List of Bacterial Species
bioMérieux has long been a partner to healthcare professionals in the battle against AMR and HAIs in particular - offering innovative culture media, ID/AST reagents and instruments for rapid, reliable microbiology testing along with environmental control solutions and educational support.
With EPISEQ® CS, we are now able to offer you an application that will help you manage HAIs in your facilities. The online tool puts NGS technology, along with one of the world's most extensive databases of bacterial strains at your disposal. You will gain critical information to track, help prevent, contain, and stop the spread of infectious diseases and HAIs.
bioMérieux has identified 14 bacterial species that cover most HAI-related organisms:
- Acinetobacter baumannii
- Clostridioides difficile
- Enterococcus faecalis
- Enterobacter cloacae complex
- Klebsiella oxytoca
- Staphylococcus aureus
- Burkholderia cepacia complex
- Escherichia coli / Shigella
- Klebsiella pneumoniae
- Enterococcus faecium
- Citrobacter freundii
- Pseudomonas aeruginosa
- Klebsiella aerogenes
- Serratia marcescens
EPISEQ® 16S
NOT FOR DIAGNOSTIC USE
For Microbiome Profiling Studies
EPISEQ® 16S helps to investigate the role of MICROBIOME on patient's health. It covers the full diversity of bacteria and archaea with relative abondance and richness. This solution supports users to discover predictive biomarkers within microbiota.
Ease of Use
- An intuitive and easy solution to identify patterns in microbiota and their associations with clinical outomes or the impact of antibiotic treatment on a given microbiota
- Directly import Illumina 16S amplicon raw data (FASTQ)
- Extensive set of visualization tools to explore data
- Built-in statistics that allow you to trust your results
- Time-interaction analysis
- Customized report that displays what is important for you to know
Microbiology Expertise / Antimicrobial Resistance
- Covers the full diversity of bacteria and archaea (SILVA 138, 2019-12-16)
- Display of relative abundance
- Display of alpha and beta diversity
- Clustering of data
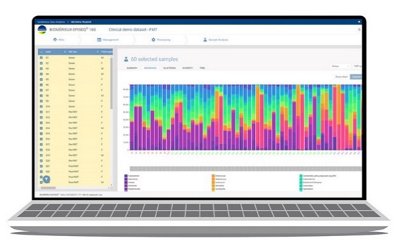
EPISEQ® 16S
Fast and Intuitive Workflow
01 – Upload
From your personal account you can directly upload your Illumina FASTQ files.
(Min. coverage recommended: 100k reads per sample; Min. sequencing length: 300 PE)
02 – Analyze
Using a straightforward subscription model, the application automatically runs all necessary steps, starting from denoising and quality assessment of raw Illumina sequencing data, over characterization of the microbiome composition of each sample, to further user-interactive visualization and statistical analyses.
EPISEQ® 16S allows you to perform microbiome time-series analysis to provide critical insights regarding potential trends changes into the structure and function of microbial communities over time.
03 – Report
A report is available once the analysis is complete. The report can be customized to include only the graphs and results you want to see included.
EPISEQ® SARS-COV-2
NOT FOR DIAGNOSTIC USE
For SARS-CoV-2 Variant Identification
EPISEQ® SARS-COV-2 allows to identify SARS -COV-2 genomic variants without bioinformatics skills. Users can perform on routine base variant analysis and download the required files to submit to public health authority for compliance (FastA file, variant call and BAM files).
Ease of Use
- A one-click solution to identify SARS-CoV-2 variants starting from raw sequencing data
- Direct import of FASTQ files:
- Illumina (paired-end or single-end)
- Thermo Fisher Ion Torrent™
- Oxford Nanopore Technologies
- A comprehensive report per sample is available for download
Microbiology Expertise / Antimicrobial Resistance
- Display of quality control information
- Variant identification based on Nextstrain¹ and Pango² nomenclature
- Weekly updates of the Pango and Nextstrain nomenclatures
- List of mutations detected in all genes of the SARS-CoV-2 genome, including the spike gene
- Identification of Variant of Concern based on WHO³ and CDC⁴
- Alignment of reads against the reference available for download in BAM format
- Genome assembly available for download in FASTA format
Fast and Intuitive Workflow
01 – Upload
From your personal account you can directly upload your:
- Illumina (paired-end or single-end)
- Thermo Fisher Ion Torrent™
- Oxford Nanopore Technologies FASTQ files
Use of targeted amplicon-based or targeted capture-based approaches are recommended.
Compatible primer sets:
- ARTIC v3
- ARTIC v4
- ARTIC v4.1
- VarSkip Short (VSS)
- Midnight (v1200)
- QIASeq DIRECT
02 – Analyze
Using a straightforward subscription model, you will get access to the cloud service that performs all steps necessary, starting from input validation, followed by quality assessment of the raw sequencing data, an alignment mapping of the data against the reference genome, a genome assembly, variant identification, and mutation screening.
03 – Report
A sample pdf report is available once the analysis is complete.
The report can be consulted in the EPISEQ® SARS-COV-2 application or downloaded if desired.